ProteoBench is an open platform for benchmarking proteomics data analysis workflows.
There several modules each one focusing on a specific Protomics technic:
- DDA quantification ion-level
- DIA quantification
and several others currently in development.
The DDA quantification module aims at comparing the capacity of Proteomics workflows to correctly estimate the intensities of ion currents. The datasets contains 6 MS runs. It is composed of a mix of different species (H. Sapiens, S. Cerevisiae, E. Coli) in known relative quantities.
- 3 replicates for condition A
- 3 replicates for condition B
Relative proportions of species (details available here):
| Sample | H. Sapiens | S. Cerevisiae | E. Coli |
|---|---|---|---|
| A | 1 | 2 | 1 |
| B | 1 | 1 | 4 |
Quantification results are available for several software (including i2MassChroQ) allowing comparison between them.
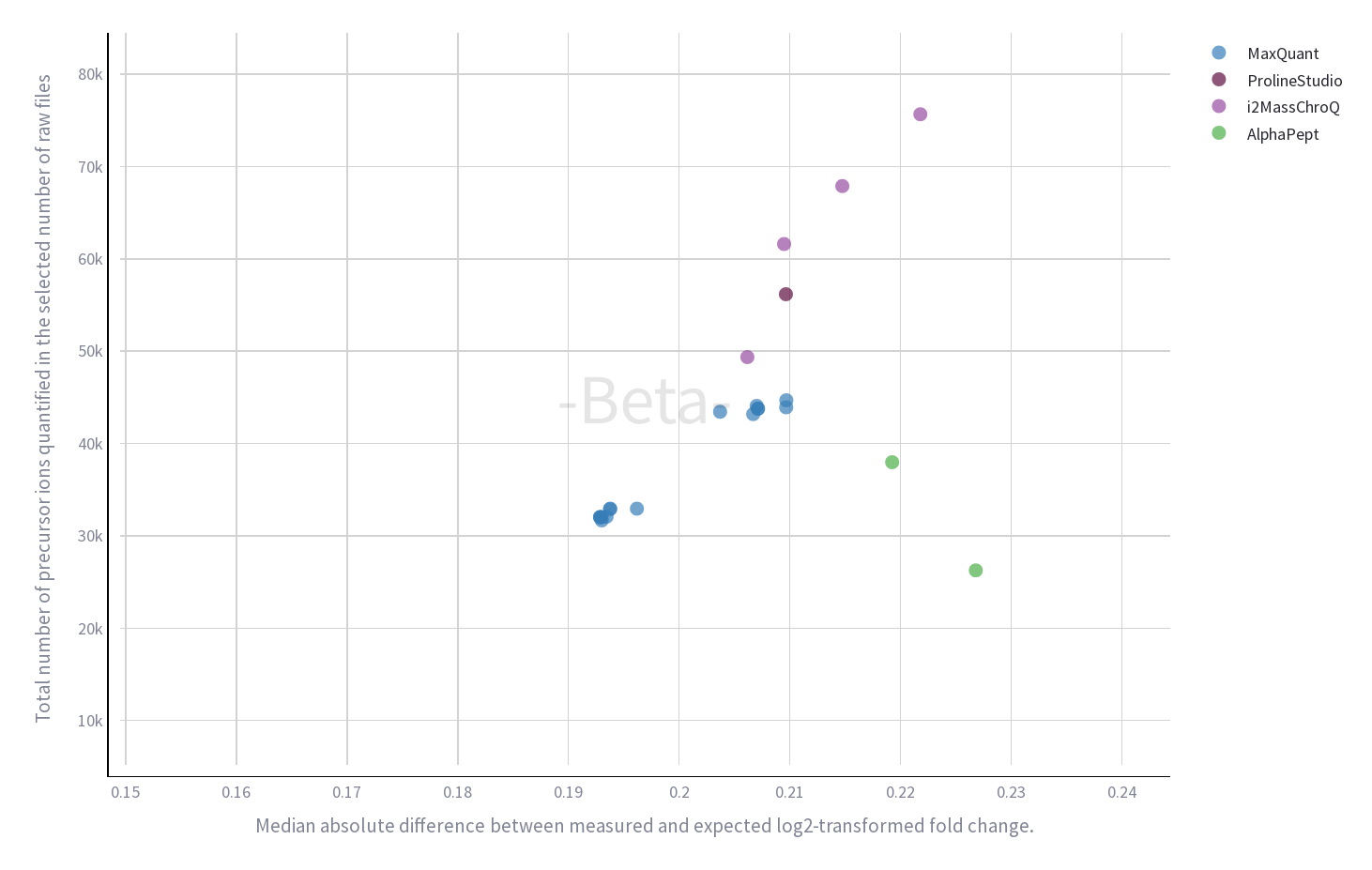
i2MassChroQ demonstrates good efficiency to extract and quantify peptides at ion level compared to MaxQuant, ProlineStudio and AlphaPept while keeping a good ratio estimation.
The results are similar to those published in :