Some changes since the last announced version 0.4.27 :
- Better timsTOF pro data management : performance enhancements, user interface and data exports (1/K0 mobility windows, collision energy)
- MassChroQ 2.4.3 integration, direct peptides quantification on timsTOF raw data.
- Direct peptide/protein identification of timsTOF runs with X!Tandem.
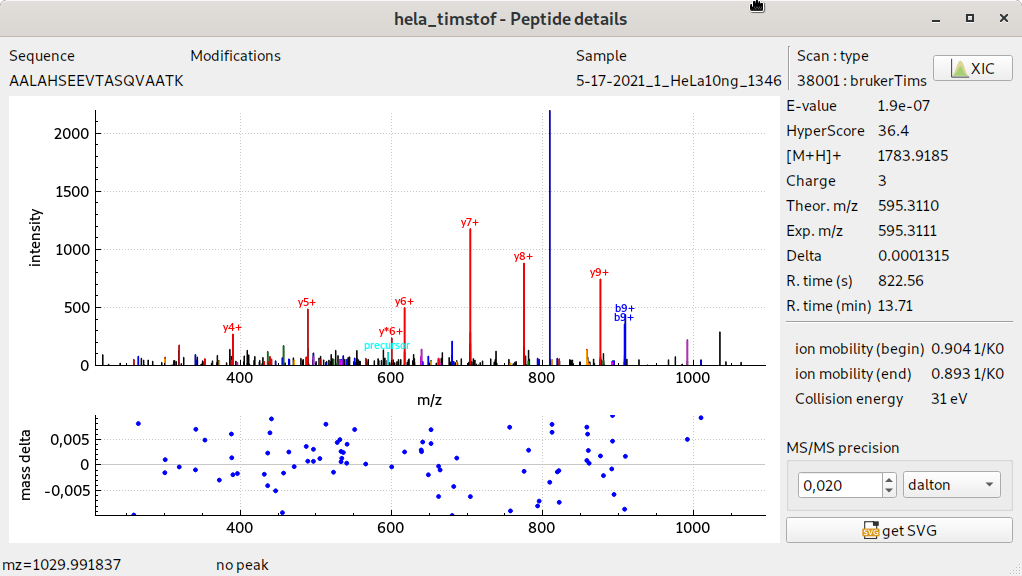
MS/MS timsTOF pro spectrum display
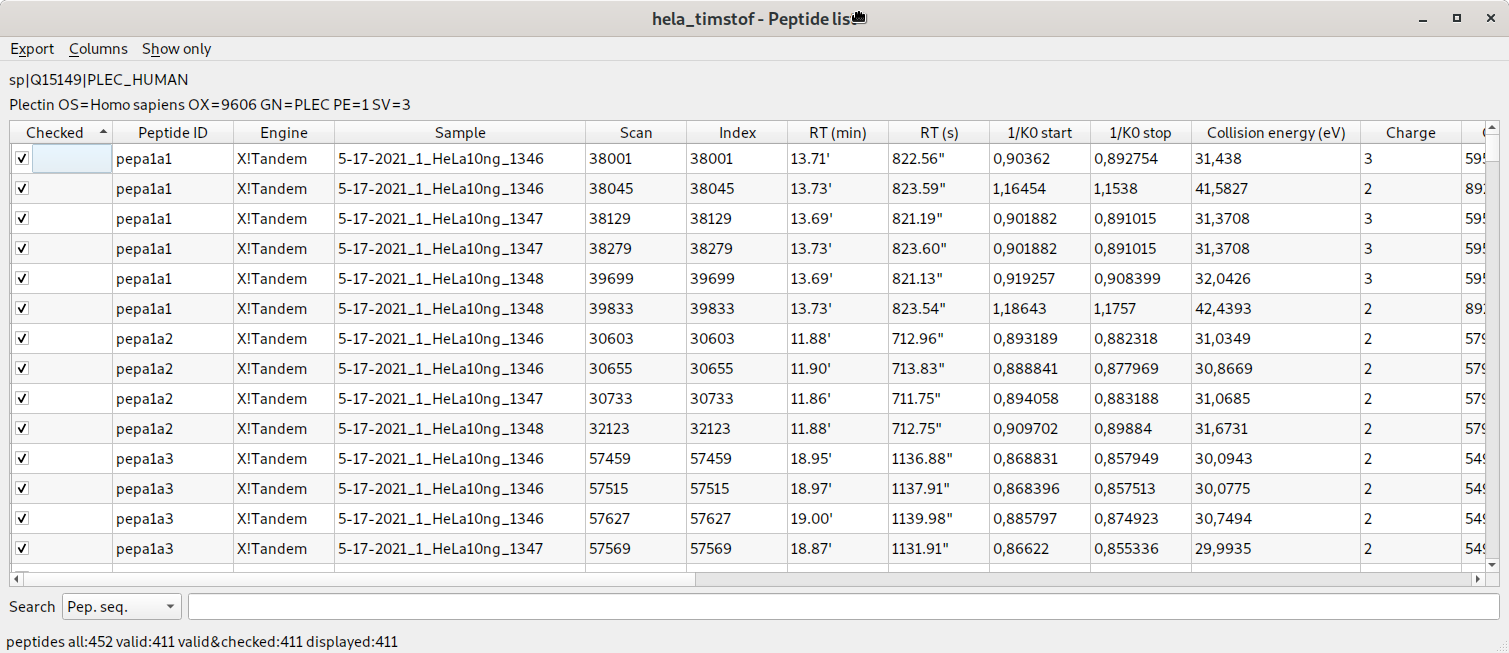
ion mobility informations displayed in the peptide list
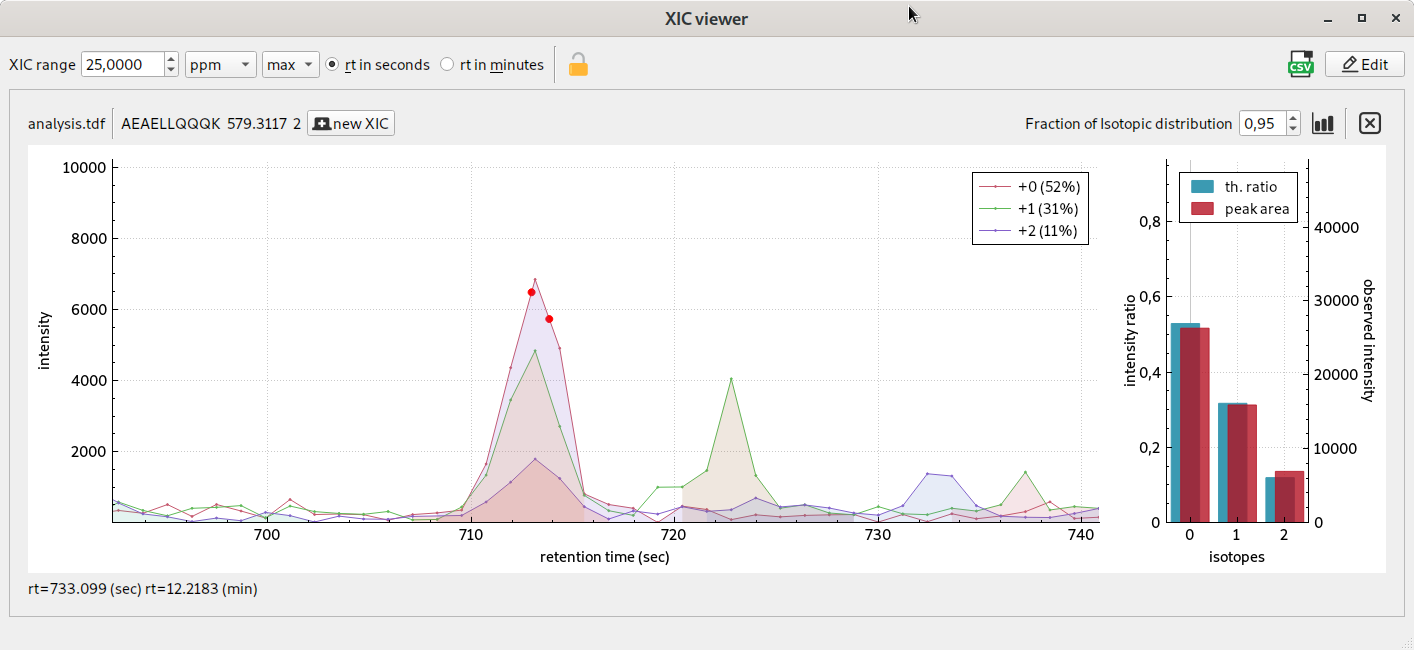
XIC viewer using mobility layer
Thanks to the whole PAPPSO team and specially to :
- Filippo Rusconi, for the documentation and C++ development.
- Thomas Renne, Bioinformatics trainee who has developped the new X!TandemPipeline features during the last 2 years.
A new post treatment module will be integrated soon to allow protein abundance computation from peptide intensities, data mining, filters, imputation, protein abundance variation analyses for different scenarii: label free, frationation, isotope labeling.
Any contribution is welcome, as well as suggestions and report of issues : ForgeMIA gitlab