PROTÉOME PEPTIDOME VERT days, the 1rst and 2th of june 2026 at Grenoble Alpes University
PROTÉOME VERT Days on June 1 and 2, 2026
Bâtiment IMAG, Université Grenoble Alpes, 150 place du Torrent, 38400 Saint-Martin-d’Hères
Bâtiment IMAG, Université Grenoble Alpes, 150 place du Torrent, 38400 Saint-Martin-d’Hères
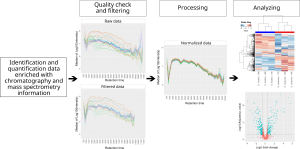
MCQR is an R package R package for the in-depth exploration, processing, and analysis of quantitative proteomics data generated from either data-dependent or data-independent acquisition methods. It has been recently accepted for publication in Journal of Proteome research
INRAE, 27 rue Sully, Dijon
The Green Proteome Days are open to people interested in plant proteomics. You are invited to present new methodological developments in the fields of sample preparation, mass spectrometry and bioinformatics analysis as well as biological results obtained with proteomics. The conference is organized by the Green Proteome network.
The agroecological transition is a major challenge for more sustainable and environmentally friendly agriculture. To contribute to this, it is essential to better understand how agroecosystems function, through interdisciplinary research that takes into account the genetic diversity of living organisms and their multiple interactions in situ, such as interactions between neighboring plants or between plants and the soil in open fields. This research is generally based on the integration of multi-omic data acquired from a very large number of samples. In this context, proteomics is becoming one of the molecular phenotyping tools offering new opportunities in agroecology.
Following an expansion process aimed at integrating new expertise, the ProFI national proteomics infrastructure will incorporate the PAPPSO platform on January 1, 2026.