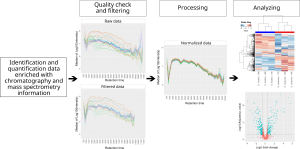
MCQR is an R package R package for the in-depth exploration, processing, and analysis of quantitative proteomics data generated from either data-dependent or data-independent acquisition methods. It has been recently accepted for publication in Journal of Proteome research
The strengths of MCQR are the following:
- Originality: MCQR leverages experimental retention time measurements for quality control, data filtering, and processing. It also facilitates the examination of interaction effects in multifactorial experiments by automating the writing of contrasts directly from the ANOVA model and performing differential analyses accordingly.
- Flexibility: The modular architecture of MCQR accommodate various types of proteomics experiments, including label-free, label-based, fractionated, or those enriched for specific post-translational modifications.
- User-friendlyness: MCQR functions, designed as simple building blocks, make it easy to test parameters and methods, and to construct customized analysis scenarios.
The source code of MCQR is released under the GPL license and can be downloaded from https://forgemia.inra.fr/pappso/mcqr. A user manual, a tutorial, and the command lines for installation on Windows, Debian, and Ubuntu are available at the same address.