MassChroQ User Manual
1 Generalities
In this chapter, some general concepts around the MassChroQ program and the reference to be used to cite the software in publications will be introduced.
1.1 General concepts and terminologies #
This section describes the general concepts at the basis of the analysis of quantitative proteomics data that one needs to grok in order to properly assimilate the workings of the MassChroQ software.
1.1.1 Spectral Counts or Area Under the Curve? #
MassChroQ is a program to perform quantitative mass spectrometry experiments in the context of proteomics projects. Quantitative proteomics projects can be handled in various ways, depending on the data that are used to actually quantify the identified peptides and proteins.
There are two main methods to peform quantitative assessments starting from proteomics data: spectral counting computations and area under the curve computations, as described below.
Spectral counts quantitation: this variant is based on the counting of the mass spectra that have allowed the identification of a given protein. It has the advantage of being a label-free quantitative technique, but it is also too rough to be considered a “robust and precise” technique.
Area under the curve quantitation: this variant is based on the integration of the ion current that can be extracted from the mass data (“extracted ion current”, or XIC) for the precursors ions that, upon fragmentation, led to the protein identification. By integrating the XICs for all the peptide precursors that identified a given protein, the software can estimate the abundance of that specific protein. The software thus needs to be able to extract the ion current for all the precursor ions in the MS spectra, when these ions were fragmented and allowed identifying peptides that in turn identified proteins. This is the process that MassChroQ implements, which is why it is called Mass Chromatogram Quantification, with Chromatogram referring to the extracted ion current (XIC) chromatogram used to assign an intensity value to the peptide ion that was fragmented.
Note
It should be noted that MassChroQ does not know anything about proteins. Instead, it does know about a list of peptidic precursor ions' m/z values, which is all it needs to perform these XIC extractions.
1.1.2 Outline of a MassChroQ working session #
The very first step is to produce the masschroqml file that lists all the peptides
that need a XIC extraction. There are two possibilities for this:
X!TandemPipeline can produce that file and has a MassChroQ configuration interface that is well documented in the user manual. The
masschroqmlfile contains all the data required for the MassChroQ operation.For scientists not using X!TandemPipeline for their proteomics projects, it is possible to craft the
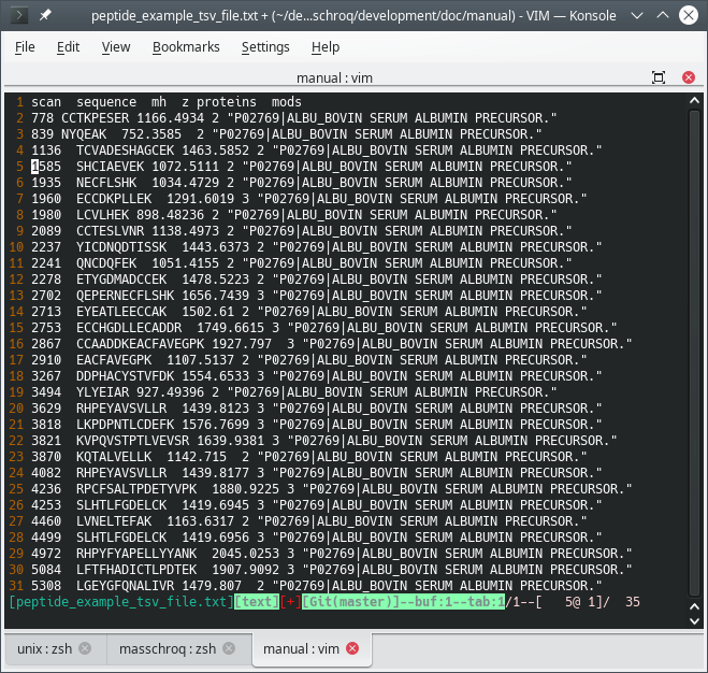
masschroqmlfile starting from a rudimentary file and by pointing MassChroQ to atsvfile (a tab-separated values file) containing the data required by MassChroQ. A typical file is shown in Figure 1.1, “The peptide list file intsvformat”.
Note
It is important that the mh column be filled-in with the calculated [M+H]+ ion mass. The charge column (z column) represents the actual charge of the precursor peptide ion.
Figure 1.1: The peptide list file in tsv format #
FIXME: brief description of the steps of the quantification analysis.
Then do this: FIXME
1.2 Citing the MassChroQ software. #
Please, cite the software using the following reference:
Benoît Valot, Olivier Langella, Edlira Nano, Michel Zivy. 2011. MassChroQ: A versatile tool for mass spectrometry quantification. Proteomics, 11: 3572-3577. https://doi.org/10.1002/pmic.201100120.
1.3 Installation of the software #
The installation material is available at http://pappso.inrae.fr/en/bioinfo/masschroq/download/.
1.3.1 Installation on MS Windows #
The installation of the software is extremely easy on the MS-Windows platform: the installation program is standard and requires no explanation.
1.3.2 Installation on Debian- and Ubuntu-based systems #
The installation on Debian- and Ubuntu-based GNU/Linux platforms is also extremely easy (even more than in the above situations). MassChroQ is indeed packaged and released in the official distribution repositories of these distributions and the only command to run to install it is:
$
[2]
sudo apt install <package_name>RETURN
In the command above, the typical package_name is
in the form masschroq for the program package and
masschroq-doc for the user manual package.
Once the package has been installed, the program shows up in the Science menu. It can also be launched from the shell using the following command:
$ masschroqRETURN